Molecular and Cytogenetic Studies in Hematology

Illustration of DNA strands and chromosomes highlighting the integration of molecular diagnostics and cytogenetic analysis in molecular and cytogenetic studies in hematology.
Molecular and cytogenetic studies play a central role in modern hematology, providing precise diagnostic, prognostic, and therapeutic insights across a wide spectrum of blood disorders. Advances in next-generation sequencing (NGS), PCR-based assays, fluorescence in situ hybridization (FISH), and karyotyping have transformed the evaluation of leukemias, lymphomas, myeloproliferative neoplasms, and inherited hematologic conditions. These techniques allow clinicians to detect actionable mutations, chromosomal abnormalities, clonal markers, and treatment-defining genetic signatures with high accuracy. As precision medicine continues to evolve, understanding the principles, applications, and clinical impact of molecular and cytogenetic testing has become essential for delivering optimal, individualized care in hematology.
Difference Between Cytogenetics and Molecular Genetics:
Molecular and cytogenetic studies are complementary diagnostic tools in hematology, each providing distinct levels of genetic insight. Cytogenetic analysis—including conventional karyotyping and fluorescence in situ hybridization (FISH)—examines chromosomal structure, number, and large-scale abnormalities such as translocations, deletions, and aneuploidy. These techniques are essential for identifying hallmark cytogenetic lesions in leukemias, lymphomas, and myeloproliferative neoplasms. In contrast, molecular testing focuses on detecting specific gene mutations, small insertions or deletions, copy-number variations, and clonal markers at the DNA or RNA level using methods such as PCR, RT-PCR, Sanger sequencing, and next-generation sequencing (NGS). While cytogenetics provides a broad overview of chromosomal architecture, molecular techniques offer high-resolution identification of actionable mutations that guide targeted therapy, risk stratification, and personalized treatment planning. Together, they form the foundation of precision medicine in modern hematology.
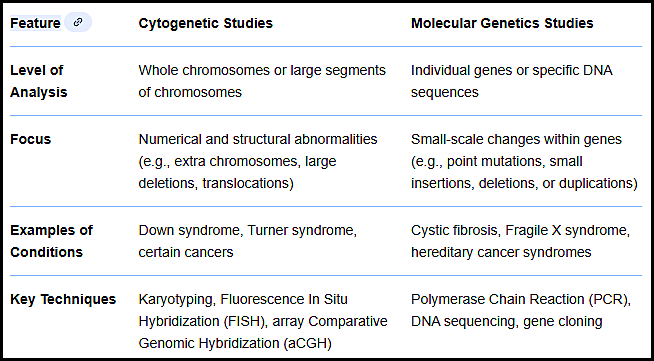
Table comparing cytogenetic and molecular genetic studies, outlining their levels of analysis, diagnostic focus, clinical applications, and key laboratory techniques in molecular and cytogenetic studies in hematology.
This review outlines the evolving landscape of cytogenetic and molecular genetic methods in the genetic characterization of hematologic malignancies. It emphasises how these methods underpin diagnosis, risk stratification, minimal residual disease detection, and targeted therapy decisions.
Cytogenetic Methods:
Cytogenetic methods are widely used during diagnosis in many types of hematological
malignancies. Classical methods like karyotyping using GTG banding technique and
molecular methods like fluorescent in situ hybridization (FISH) are still gold standard in
clinics all over the world. According to WHO 2008 classification the cytogenetic analysis is
the basic diagnostic tool during the diagnosis of blood neoplasms. Chromosomal
abnormalities have established and known prognostic significance. According to the
European Leukemia Net-Workpackage Cytogenetics – the cytogenetic diagnostic is essential for disease classification, prognostic assessment and treatment decisions. Despite the fact that new molecular methods are very promising the classical cytogenetic methods together with FISH are still the reference test in many hematological neoplasms.
Common Cytogenetic Methods:
Karyotyping (GTG banding)
This is a classical method that arranges chromosomes from a blood or bone marrow sample into pairs, allowing for the visualization of the overall structure, number, and any large-scale abnormalities like translocations, deletions, or duplications.
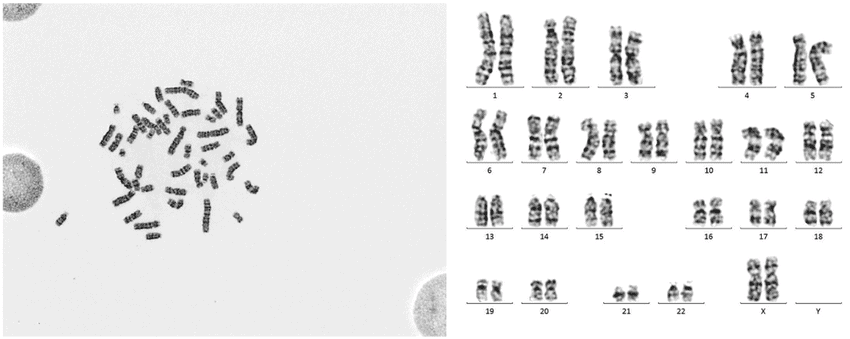
G-banded metaphase spread with corresponding karyotype used for cytogenetic evaluation of chromosomal abnormalities in hematologic disorders in molecular and cytogenetic studies in hematology.
Fluorescence In Situ Hybridization (FISH)
This technique uses fluorescent probes that bind to specific DNA sequences on chromosomes. It’s particularly useful for identifying specific gene fusions, deletions, or translocations that may be too small to see with a standard karyotype, and can be used when cells don’t divide well enough for karyotyping, such as in multiple myeloma.
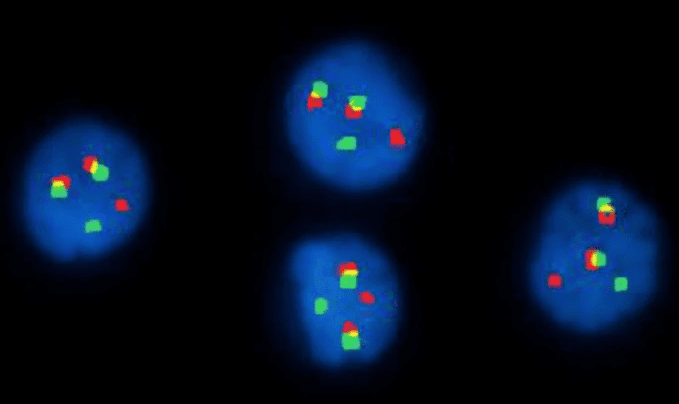
Interphase FISH with IGH/FGFR3 dual-fusion probes showing the t(4;14) translocation in multiple myeloma, a key finding in molecular and cytogenetic studies in hematology.
Spectral Karyotyping (SKY) / Multicolor FISH (mFISH)
This technique uses multiple differently colored fluorescent probes to visualize each chromosome pair in a unique color, allowing precise identification of complex or cryptic chromosomal rearrangements. It is particularly useful in cases with highly abnormal karyotypes, such as advanced leukemias or secondary MDS.
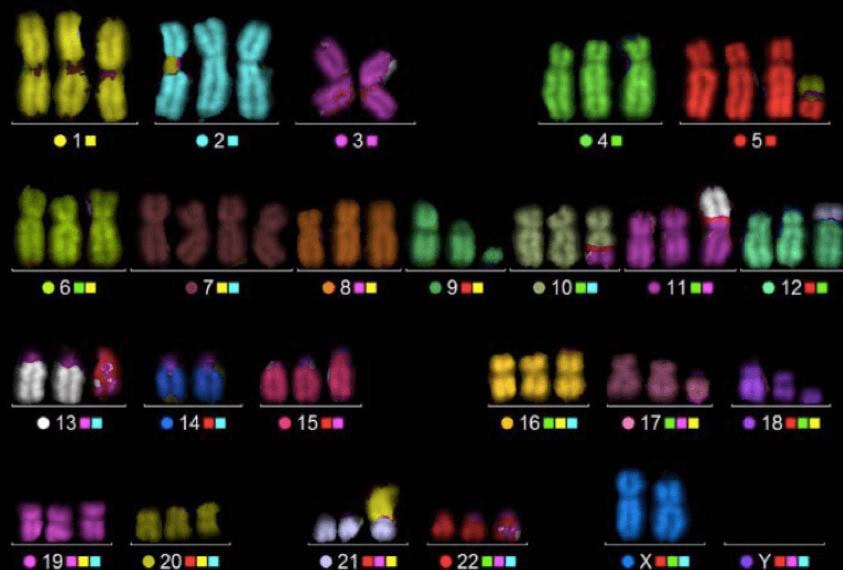
Multicolor FISH (M-FISH) karyotype with each chromosome pair assigned a unique spectral color, enabling precise identification of complex or cryptic chromosomal rearrangements in molecular and cytogenetic studies in hematology.
Telomere and Centromere FISH Probing
Targeted probes designed for chromosome ends (telomeres) or centromeres help identify subtle terminal deletions, ring chromosomes, or mis-segregation events, which are common in bone marrow failure syndromes and certain leukemias.
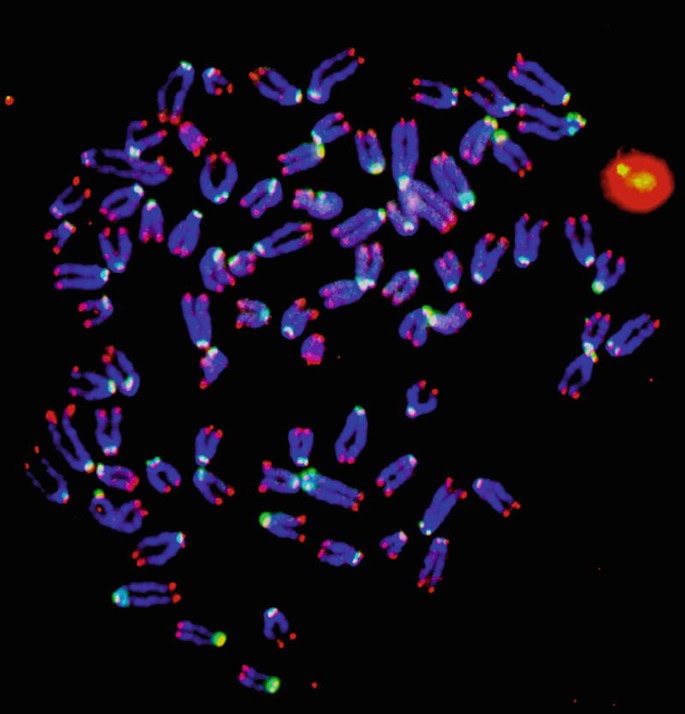
Telomere and centromere FISH staining highlighting fluorescent chromosome ends and centromeric regions used in molecular and cytogenetic studies in hematology.
Microarray analysis
This method is used to detect very small deletions or duplications of chromosome segments, known as copy number variations. It can identify abnormalities that are too small to be detected by karyotyping alone.
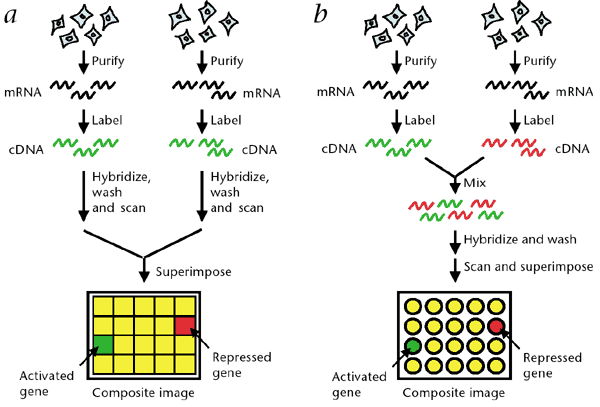
Two microarray gene expression workflows comparing separate and dual-label hybridization to detect activated and repressed genes in molecular and cytogenetic studies in hematology.
Array Comparative Genomic Hybridization (aCGH)
While related to microarray analysis, aCGH specifically compares patient DNA to reference DNA, enabling high-resolution detection of copy-number gains and losses across the entire genome. It can uncover submicroscopic abnormalities undetectable by karyotype or routine FISH.
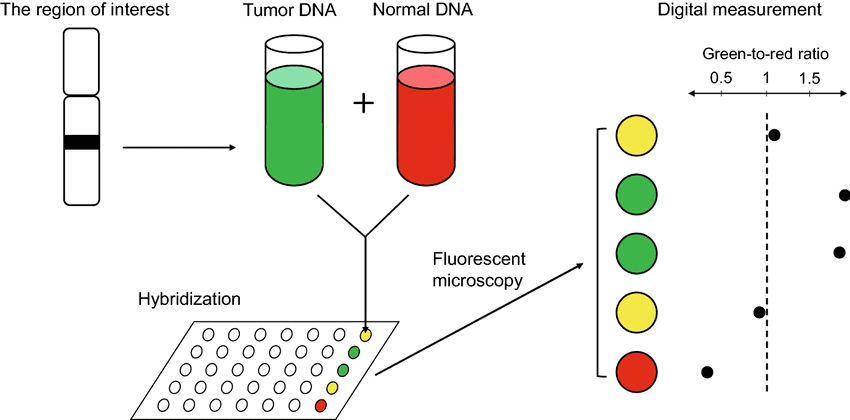
Array CGH workflow illustrating tumor and normal DNA hybridization used in molecular and cytogenetic studies in hematology for detecting genomic gains and losses.
Single Nucleotide Polymorphism (SNP) Array
SNP arrays detect both copy-number changes and regions of copy-neutral loss of heterozygosity (LOH), which are clinically relevant in disorders like CLL, MDS, and AML. LOH can indicate acquired uniparental disomy affecting tumor suppressor genes.
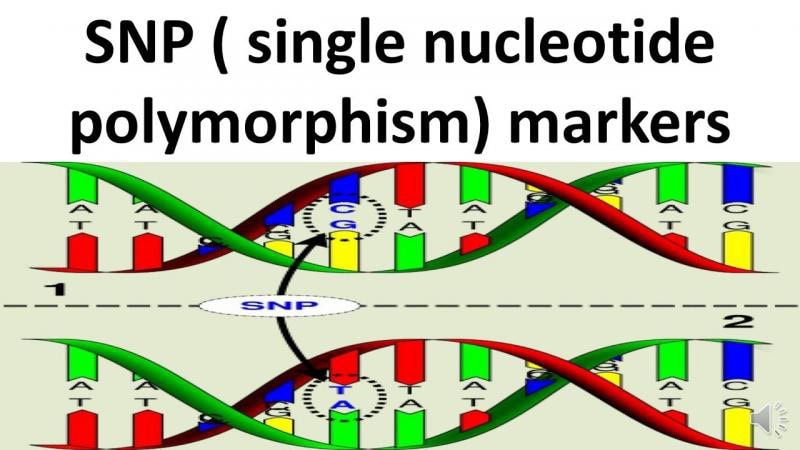
Illustration of single nucleotide polymorphism (SNP) markers showing DNA base substitutions used in molecular and cytogenetic studies in hematology.
Hematologic Impact of Key Cytogenetic Methods:
Commonly used cytogenetic methods have significant diagnostic, prognostic, and therapeutic implications across hematologic malignancies. Conventional karyotyping remains essential for identifying hallmark chromosomal abnormalities such as t(8;21), inv(16), t(15;17), del(5q), −7, and +8, all of which directly influence risk stratification and treatment pathways in AML and MDS. Interphase FISH is vital in disorders where metaphase spreads are difficult to obtain, particularly in multiple myeloma, allowing rapid detection of high-risk lesions such as t(4;14), del(17p), and 1q gain. SNP arrays and aCGH complement traditional methods by revealing submicroscopic copy-number changes and copy-neutral loss of heterozygosity, refining diagnoses in CLL, MDS, and myeloproliferative neoplasms. Together, these cytogenetic tools provide a comprehensive genomic profile that guides targeted therapy, informs transplant decisions, and predicts disease behavior, underscoring their central role in modern hematologic practice.
Molecular Methods:
Molecular testing has become an indispensable component of modern hematologic diagnostics, providing high-resolution insights into gene mutations, fusion transcripts, clonal markers, and treatment-defining genomic signatures. These methods complement cytogenetics by detecting submicroscopic abnormalities, quantifying disease burden, guiding targeted therapy, and refining prognostic models across leukemias, lymphomas, myeloproliferative neoplasms (MPNs), myelodysplastic syndromes (MDS), and inherited hematologic conditions. Below are the most widely used and clinically relevant molecular techniques in routine hematology practice.
Common Molecular Methods:
Polymerase Chain Reaction (PCR)–Based Techniques
*Conventional PCR:
Used for rapid detection of known mutations or small genetic alterations.
Applications:
- JAK2 V617F mutation in myeloproliferative neoplasms (MPNs)
- BCR–ABL1 fusion screening
- MYD88 L265P in lymphoplasmacytic lymphoma
Advantages:
Fast, inexpensive, highly sensitive.
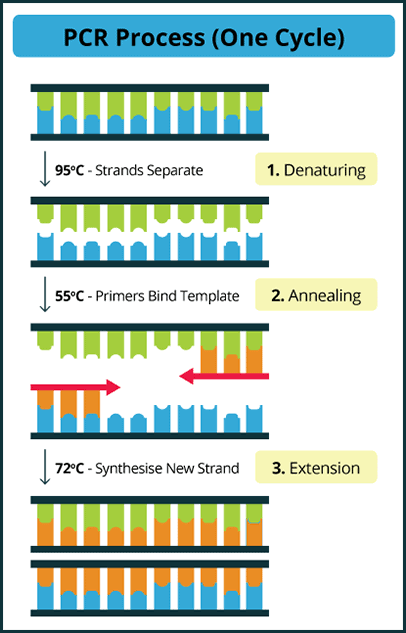
Diagram of the PCR cycle showing denaturation, annealing, and extension steps used in molecular and cytogenetic studies in hematology.
*Real-Time Quantitative PCR (qPCR):
Measures gene targets in real time with fluorescent probes.
Applications:
- Quantitative BCR–ABL1 monitoring in CML
- FLT3-ITD allelic ratio in AML
- MRD assessment in ALL and AML
This remains the gold standard for measurable residual disease assessments in many hematologic cancers.
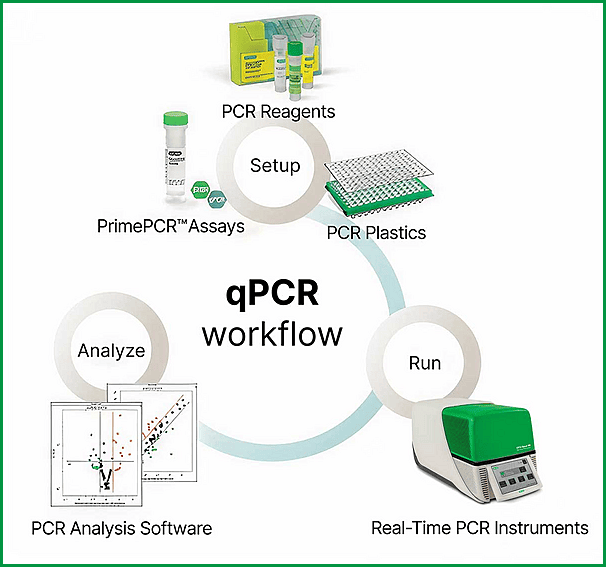
qPCR workflow illustrating setup, amplification, and data analysis steps used in molecular and cytogenetic studies in hematology.
*Reverse Transcription PCR (RT-PCR):
Detects fusion transcripts or gene expression at the RNA level.
Examples:
- PML–RARA in acute promyelocytic leukemia
- ETV6–RUNX1 in pediatric ALL
- BCR–ABL1 p210 vs. p190 transcript typing
*Digital PCR (dPCR)
An emerging, ultra-sensitive method that partitions reactions into thousands of droplets for precise quantification.
Clinical Strengths:
- MRD monitoring in AML, ALL, and CML
- Detection of low-level TP53 mutations
- Monitoring JAK2 V617F allele burden
dPCR often outperforms qPCR in sensitivity and reproducibility.
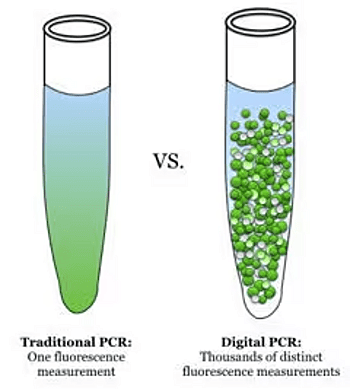
Comparison of traditional PCR and digital PCR illustrating single versus thousands of fluorescence measurements used in molecular and cytogenetic studies in hematology.
Sanger Sequencing
The classical sequencing method for detecting single-gene mutations or hotspot variants.
Still used for:
- Confirming JAK2, CALR, or MPL mutations
- Verifying NPM1 mutations
- Small gene panels where NGS is unnecessary
Although replaced by NGS in many settings, Sanger remains reliable for confirmation and targeted mutation analysis.
Next-Generation Sequencing (NGS)
NGS allows simultaneous interrogation of dozens to hundreds of genes, offering broad mutation profiles critical for contemporary hematologic practice.
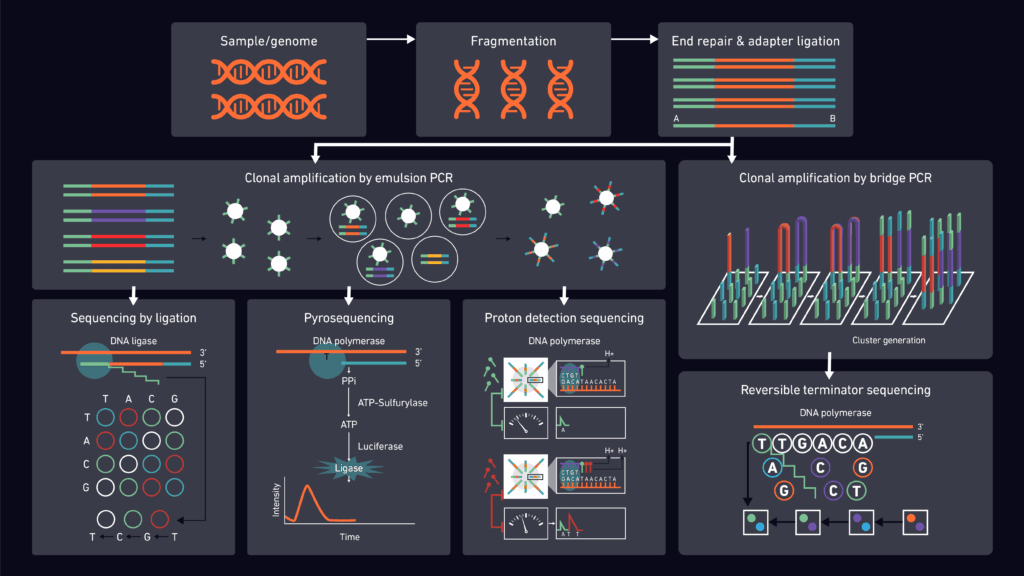
Next-generation sequencing (NGS) workflow illustrating DNA preparation, clonal amplification, and multiple sequencing chemistries used in molecular and cytogenetic studies in hematology.
*Targeted Gene Panels
Most commonly used in clinical hematology.
Detects:
- Point mutations
- Small insertions/deletions
- Copy-number variants
- Some fusions (depending on platform)
Applications:
- AML: FLT3, NPM1, IDH1/2, ASXL1, DNMT3A
- MDS: SF3B1, TP53, TET2, SRSF2
- CLL: TP53, NOTCH1, SF3B1
- MPNs: JAK2, CALR, MPL plus additional co-mutations
These mutations guide prognosis, treatment selection, transplant decisions, and clinical trial eligibility.

Venn diagram comparing genes covered in hematologic malignancy and solid tumor NGS panels, highlighting shared targets important in molecular and cytogenetic studies in hematology.
*RNA Sequencing (RNA-seq)
Used to detect gene fusions, aberrant transcripts, splicing abnormalities, and expression signatures.
Value in hematology:
- Identifying cryptic or novel fusion genes
- Classifying ambiguous leukemias
- Research-level profiling of tumor microenvironment
*Whole Exome Sequencing (WES) / Whole Genome Sequencing (WGS)
Used mainly in academic settings or complex diagnostic cases.
Strengths:
- Comprehensive detection of coding variants (WES)
- Genome-wide detection of structural variants, insertions, deletions (WGS)
Clinical niche:
- Congenital bone marrow failure syndromes
- Unexplained cytopenias or inherited predisposition to hematologic malignancy
Fragment Analysis (Capillary Electrophoresis)
Measures length variations in PCR-amplified DNA fragments.
Common uses:
- FLT3-ITD detection in AML
- T-cell receptor (TCR) and immunoglobulin (IGH) clonality
- Microsatellite instability (rarely in hematology)
- Chimerism analysis post-transplant
It remains a staple in molecular hematopathology labs.
Next-Generation Minimal Residual Disease (MRD) Assays
Highly sensitive assays using NGS, digital PCR, or allele-specific techniques.
Clinical role:
- Deep MRD quantification in AML, ALL, CLL, and myeloma
- Predicting relapse risk
- Guiding post-remission therapy and transplant decisions
MRD-directed treatment is now incorporated into multiple international guidelines.
Flow Cytometric Immunophenotyping (Molecular-Adjacent)
Although not a molecular technique per se, flow cytometry provides complementary clonal and phenotypic data that supports molecular findings, especially in MRD.
Emerging Molecular Technologies
*Long-Read Sequencing (PacBio, Oxford Nanopore)
Helps identify complex structural variants and long-range genomic changes.
*Single-Cell Genomics
Dissects clonal evolution, tumor heterogeneity, and rare subclones.
*Cell-Free DNA (cfDNA) / Liquid Biopsy
An increasingly valuable tool in lymphomas, AML, and post-transplant monitoring.
Comprehensive Cytogenetic and Molecular Testing Guide in Hematology:
Key Cytogenetic & Molecular Abnormalities by Hematologic Malignancy
| Hematologic Malignancy | Key Cytogenetic Abnormalities | Key Molecular Mutations | Notes |
|---|---|---|---|
| AML | t(8;21), inv(16), t(15;17), +8, −7, del(5q), complex karyotype | FLT3-ITD/TKD, NPM1, CEBPA, IDH1/2, TP53, ASXL1, DNMT3A | ELN risk heavily genetics-driven |
| ALL (B-ALL) | t(12;21), t(9;22), hyperdiploidy, t(1;19) | IKZF1 deletion, CRLF2, JAK pathway | MRD crucial; fusion-driven subtypes |
| ALL (T-ALL) | TCR locus rearrangements, del(6q) | NOTCH1, FBXW7, PTEN | Genetics guide prognosis |
| CLL | del(13q), trisomy 12, del(11q), del(17p) | TP53, NOTCH1, SF3B1, BIRC3 | IGHV mutation status critical |
| CML | t(9;22) BCR-ABL1 | BCR-ABL1 transcript variants | qPCR for monitoring |
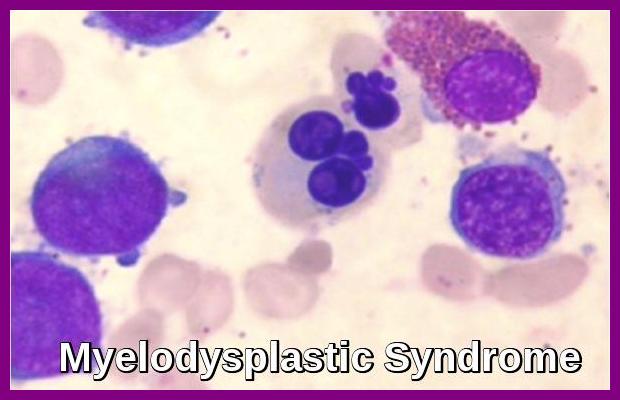
| MDS | del(5q), −7, +8, complex karyotype | SF3B1, TET2, TP53, ASXL1, SRSF2 | IPSS-R relies heavily on genetics |
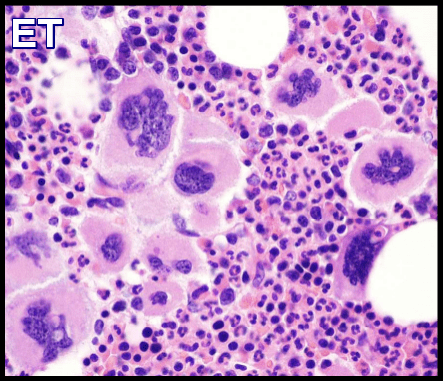
| MPNs (PV/ET/MF) | Usually normal; occasional +9, +8 | JAK2, CALR, MPL | Driver mutations nearly diagnostic |
| Multiple Myeloma | t(11;14), t(4;14), t(14;16), +1q, del(17p), del(13q) | NRAS, KRAS, BRAF, TP53 | iFISH standard for risk |
| Lymphoplasmacytic Lymphoma | del(6q) | MYD88 L265P, CXCR4 | MYD88 hallmark mutation |
| Mantle Cell Lymphoma | t(11;14) CCND1-IGH | ATM, TP53, NOTCH1 | SOX11 helps classification |
| DLBCL | MYC, BCL2, BCL6 rearrangements | MYD88, EZH2, CD79B | “Double hit” = MYC + BCL2/BCL6 |
| Follicular Lymphoma | t(14;18) BCL2-IGH | EZH2, CREBBP, KMT2D | Classic germinal-center genetics |
| Burkitt Lymphoma | MYC t(8;14) or variants | ID3, TCF3 | Very high proliferation rate |
Which Test to Order for Each Malignancy (Clinician’s Quick Guide)
| Condition | First-Line Cytogenetic Tests | Molecular Tests |
|---|---|---|
| AML | Karyotype + AML FISH panel | NGS panel (FLT3, NPM1, CEBPA, IDH1/2) |
| ALL | ALL FISH panel | PCR for BCR-ABL1; NGS for IKZF1, JAK pathway |
| CLL | CLL FISH panel | TP53 mutation; IGHV status; NGS panel |
| MDS | Karyotype | NGS (SF3B1, TP53, ASXL1, TET2); SNP array |
| CML | Karyotype for Philadelphia chromosome | qPCR for BCR-ABL1 transcript (p210/p190) |
| MPNs (PV/ET/MF) | Cytogenetics optional; occasionally +9 or +8 | JAK2 → CALR → MPL PCR/NGS |
| Multiple Myeloma | iFISH (t(4;14), del(17p), +1q) | NGS for RAS/RAF pathway, TP53 |
| Lymphoplasmacytic Lymphoma (Waldenström) | Karyotype (optional) | MYD88 L265P; CXCR4 mutations |
| Mantle Cell Lymphoma | FISH for t(11;14) CCND1-IGH | SOX11; NGS for aggressive variants |
| DLBCL / High-Grade Lymphoma | FISH for MYC / BCL2 / BCL6 (“double hit”) | NGS for MYD88, EZH2, CD79B |
| Aplastic Anemia / Bone Marrow Failure | Telomere length, chromosome breakage studies | Inherited marrow failure panel (TERT, TERC, DKC1, GATA2) |
What Test Detects What? (Simplified Exam & Teaching Guide)
| Test | Best for Detecting | Examples |
|---|---|---|
| Karyotyping (G-banding) | Large chromosomal abnormalities (big changes) | Aneuploidy, t(8;21), del(5q), monosomy 7 |
| FISH | Specific gene fusions, deletions, amplifications | t(4;14) in myeloma; MYC/BCL2/BCL6; del(17p) |
| aCGH / SNP Array | Copy-number changes too small for karyotype | Submicroscopic deletions/duplications, MDS lesions |
| PCR (Conventional) | Known single mutations or small fusions | JAK2 V617F; PML-RARA |
| qPCR | Quantifying mutations & MRD | BCR-ABL1 monitoring; NPM1 MRD |
| Digital PCR | Ultrasensitive mutation detection at very low VAF | TP53 low-level clones; rare mutations |
| NGS Panels | Multiple mutations across many genes simultaneously | Myeloid panels; lymphoid panels; MPN/CLL/MM panels |
| Whole Genome/Exome Sequencing | Comprehensive genomic landscape (research) | Inherited marrow failure, rare lymphomas |
| Flow Cytometry (Bonus) | Immunophenotyping (not genetic but essential) | CLL immunophenotype; MRD in ALL |
Turnaround Times for Molecular vs Cytogenetic Tests in Hematology:
Fast Turnaround Times: Molecular vs Cytogenetic Tests in Hematology
Molecular tests are generally much faster than cytogenetic tests. Molecular assays use extracted DNA or RNA and do not require cell culture, whereas many cytogenetic studies (especially karyotyping) depend on growing dividing cells and preparing metaphase spreads.
| Test Type | Example Tests | Typical Turnaround Time |
|---|---|---|
| Molecular | PCR, qPCR (BCR-ABL1), digital PCR, FLT3-ITD fragment analysis | ≈ 2–48 hours |
| Molecular (NGS panels) | Targeted myeloid or lymphoid NGS panels | ≈ 3–7 days |
| Cytogenetic (FISH) | Interphase FISH panels (myeloma, CLL, AML) | ≈ 1–3 days |
| Cytogenetic (Karyotype) | Conventional G-banded karyotyping | ≈ 5–14 days |
In practice: molecular tests (especially PCR and qPCR) provide the fastest results, while full cytogenetic karyotyping is slower but still essential for comprehensive risk stratification in hematologic malignancies.
Summary:
Cytogenetic testing helps detect large chromosomal abnormalities, such as aneuploidy and translocations, which are essential for diagnosing and risk-stratifying hematologic malignancies like AML, ALL, MDS, CLL, and MM. Molecular testing provides finer detail by detecting gene mutations (NPM1, FLT3, JAK2), fusion transcripts (BCR–ABL1, PML–RARA), and minimal residual disease with high sensitivity. Together, these techniques reveal both the “big picture” chromosomal landscape and the precise genetic alterations driving disease, guiding prognosis, targeted therapy, and treatment decisions.
Molecular tests (PCR, qPCR, NGS) are significantly faster than cytogenetic tests because they do not require cell culture, whereas karyotyping and many FISH analyses take several days.
Cytogenetic studies look at chromosomes. They identify big structural changes such as translocations, deletions, duplications, aneuploidy, and complex karyotypes. Molecular studies look at genes and DNA/RNA sequences. They detect small mutations, gene fusions, variant allele burden, and minimal residual disease (MRD) at much higher sensitivity.
Think of it this way:
👉 Cytogenetics = big structural map of the genome
👉 Molecular = fine details within the DNA sequence
Both approaches complement each other and are essential for modern hematology diagnostics, prognosis, and targeted therapy selection.
Questions and Answers:
What is the difference between molecular and cytogenetic testing in hematology?
Molecular testing examines DNA or RNA to detect mutations, gene fusions, and minimal residual disease, while cytogenetic testing evaluates whole chromosomes to identify structural abnormalities such as translocations, deletions, duplications, and aneuploidy. Both methods complement each other in diagnosing and risk-stratifying hematologic malignancies.
Which test gives faster results—molecular or cytogenetic testing?
Molecular tests are much faster. PCR and qPCR results return within 2–48 hours, while targeted NGS takes 3–7 days. Cytogenetic tests like karyotyping require cell culture and typically take 5–14 days, although FISH panels return results within 1–3 days.
When should clinicians order FISH instead of conventional karyotyping?
FISH is preferred when specific abnormalities are suspected, such as MYC, BCL2, BCL6, del(17p), or t(4;14), or when dividing cells are insufficient for metaphase analysis. It provides rapid, targeted detection of clinically significant genetic lesions.
Why is next-generation sequencing important in hematology?
NGS enables simultaneous analysis of multiple genes involved in hematologic malignancies, including FLT3, NPM1, TP53, ASXL1, IDH1/2, and MYD88. It enhances diagnostic precision, prognostication, targeted therapy selection, and MRD monitoring.
Why do some hematologic malignancies show normal cytogenetics but abnormal molecular results?
Cytogenetic methods detect large chromosomal abnormalities, while molecular techniques identify small mutations and submicroscopic alterations. Many leukemias and lymphomas contain gene-level mutations that are not visible by karyotyping but are easily detected by PCR or NGS.
How do genetic abnormalities influence treatment decisions in hematology?
Specific genetic lesions guide targeted therapy and prognostic classification. Mutations such as FLT3-ITD, IDH1/2, TP53, IGHV status, MYD88 L265P, and high-risk myeloma lesions like t(4;14) and del(17p) directly impact treatment strategies and long-term outcomes.
Do molecular tests detect minimal residual disease more accurately than cytogenetics?
Yes. qPCR, digital PCR, and NGS-based MRD assays can detect disease down to 10⁻⁵ or 10⁻⁶, far exceeding the resolution of cytogenetic techniques. Molecular MRD is now essential in AML, ALL, CLL, and multiple myeloma.
References:
Lancet Laboratories. Cytogenetics. 2025. Available from: https://www.lancet.co.za/cytogenetics/
Behrens YL, Pietzsch S, Antić Ž, Zhang Y, Bergmann AK. The landscape of cytogenetic and molecular genetic methods in diagnostics for hematologic neoplasia. Best Pract Res Clin Haematol. 2024;37(1):101539. doi:10.1016/j.beha.2024.101539.
Altinsoy E, Yang J, Tu E. An improved denoising of G-banding chromosome images using cascaded CNN and binary classification network. The Visual Computer. 2022;38(6):2139-2152. doi:10.1007/s00371-021-02273-5. Available from: https://www.researchgate.net/figure/G-banding-chromosome-image-and-its-karyotype_fig1_353937971 [accessed 19 Nov 2025].
Abdel-Qader HY, Fouad DA, Abuelela SA, et al. Clonal heterogeneity by fluorescence in situ hybridization in multiple myeloma: enhanced cytogenetic risk stratification. Egypt J Med Hum Genet. 2022;23:66. doi:10.1186/s43042-022-00220-0.
Stears R, Martinsky T, Schena M. Trends in microarray analysis. Nat Med. 2003;9:140–145. doi:10.1038/nm0103-140.
Mały E, Nowak J, Januszkiewicz-Lewandowska D. Cytogenetics in Hematooncology. In: Recent Trends in Cytogenetic Studies – Methodologies and Applications. InTech; 2012. Available from: https://scispace.com/pdf/cytogenetics-in-hematooncology-vkg754fnhi.pdf [accessed 19 Nov 2025].
Arber DA, Orazi A, Hasserjian R, et al. The 2016 revision to the WHO classification of myeloid neoplasms and acute leukemia. Blood. 2016;127(20):2391–2405.
Greenberg PL, Tuechler H, Schanz J, et al. Revised International Prognostic Scoring System for Myelodysplastic Syndromes (IPSS-R). Blood. 2012;120(12):2454–2465.
Creative Bioarray. Multicolor FISH (M-FISH) Analysis Service. Available from: https://www.creative-bioarray.com/services/multicolor-fish-m-fish-analysis.htm [accessed 19 Nov 2025].
Kamel YM, Mostafa Y, et al. Fluorescence in situ hybridization assays designed for del(7q) detection uncover more complex rearrangements in myeloid leukaemia cell lines. 2014.
Kantarjian H, Cortes J, O’Brien S, et al. Molecular pathogenesis and therapy of chronic myeloid leukemia. N Engl J Med. 2001;344:1031–1037.
Robust Sparse Bayesian Infinite Factor Models. Scientific Figure on ResearchGate. Available from: https://www.researchgate.net/figure/The-overview-of-array-comparative-genomic-hybridization-aCGH-procedure_fig2_359066049 [accessed 19 Nov 2025].
Cartwright IM, Haskins JS, Kato TA. PNA telomere and centromere FISH staining for accurate analysis of radiation-induced chromosomal aberrations. In: Kato T, Wilson P, eds. Radiation Cytogenetics. Methods in Molecular Biology. Vol 1984. Humana; 2019. doi:10.1007/978-1-4939-9432-8_11.
OpenPR. Single Nucleotide Polymorphism (SNP) Genotyping Market. Available from: https://www.openpr.com/news/2589026/single-nucleotide-polymorphism-snp-genotyping-market [accessed 19 Nov 2025].
Bio-Rad Laboratories. What is Real-Time PCR (qPCR)? Available from: https://www.bio-rad.com/en-ie/applications-technologies/what-real-time-pcr-qpcr?ID=LUSO4W8UU [accessed 20 Nov 2025].
Sigma-Aldrich. Dual-Labeled Probes in Digital PCR. Available from: https://www.sigmaaldrich.com/US/en/technical-documents/technical-article/genomics/pcr/dual-labeled-probes-for-digital-pcr?srsltid=AfmBOoofD9st20D9t7BBjv67S8nMzDO6qMeHundfXcv0JTj4gCMEetqq [accessed 20 Nov 2025].
Technology Networks. Next-Generation Sequencing (NGS): Definition and Overview. 2024 (Oct 28). Available from: https://www.technologynetworks.com/genomics/articles/next-generation-sequencing-ngs-definition-and-overview-346532 [accessed 20 Nov 2025].
Fox A, Hiemenz M, Lieberman D, et al. Next-generation sequencing for the detection of actionable mutations in solid and liquid tumors. J Vis Exp. 2016. doi:10.3791/52758.
Quest Diagnostics. Understanding Cytogenetic Tests vs. Molecular Genetics Tests: A Beginner’s Guide. 2023. Available from: https://www.questdiagnostics.com/our-company/actions-insights/2023-blogs/understanding-cytogenetic-tests-vs–molecular-genetics-tests–a-beginner-s-guide
American Society of Human Genetics (ASHG). What Is a Cytogenetic Test? 2019. Available from: https://www.ashg.org/genetics/ashg/education/pdf/ASHG_Cytogenetics_Primer.pdf
National Human Genome Research Institute (NHGRI). What Are Molecular Genetic Tests? 2021. Available from: https://www.genome.gov/genetics-glossary/Molecular-Genetic-Tests
National Society of Genetic Counselors (NSGC). Genetic Testing: Frequently Asked Questions. 2021. Available from: https://www.nsgc.org/p/bl/et/blogaid=310
Döhner H, Estey E, Grimwade D, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations. Blood. 2017;129(4):424–447.
Schuurhuis GJ, Heuser M, Freeman S, et al. Minimal residual disease in AML: a consensus document from the European LeukemiaNet MRD Working Party. Blood. 2018;131(12):1275–1291.
Gandhi M, Evans K, Kadia T, et al. Digital PCR in hematologic malignancies. Leukemia. 2023;37:1–12.
O’Donnell MR, Abutalib SA, Altman J, et al. NCCN Guidelines® Insights: Acute Lymphoblastic Leukemia. J Natl Compr Canc Netw. 2022;20(1):40–49.
Hallek M, et al. iwCLL guidelines for diagnosis and treatment of CLL. Blood. 2018;131(25):2745–2760.
Rajkumar SV. Multiple myeloma: 2024 update on diagnosis, risk stratification, and management. Am J Hematol. 2024;99(1):E31–E43.
Keywords:
molecular and cytogenetic studies in hematology, cytogenetic analysis in hematologic malignancies, molecular diagnostics in hematology, karyotyping in leukemia, metaphase cytogenetics hematology, G-banding chromosome analysis, interphase FISH hematology, FISH genetic abnormalities, multicolor FISH M-FISH karyotype, telomere FISH staining, centromere FISH staining, SNP array hematology, array comparative genomic hybridization aCGH, copy number variation testing hematology, chromosomal abnormalities in hematologic cancers, high-risk myeloma cytogenetics, myeloma high-risk cytogenetics t(4;14) del(17p), AML cytogenetic abnormalities, ALL molecular genetics, CLL genetic markers IGHV TP53, CLL FISH panel, MDS cytogenetics IPSS-R, myeloproliferative neoplasms molecular testing, JAK2 CALR MPL mutations, BCR-ABL1 PCR CML, IGHV mutation testing CLL, TP53 mutation hematologic malignancies, MYD88 L265P mutation, CXCR4 mutation Waldenström, NPM1 mutation AML, FLT3 ITD AML, IDH1 IDH2 mutations leukemia, CEBPA mutation AML, gene fusions in leukemia, cytogenetic complexity hematology, molecular mutation profiling hematology, digital PCR hematology, qPCR hematology, PCR workflow hematology, NGS workflow hematology, next-generation sequencing in hematology, myeloid NGS panels, lymphoid NGS panels, precision genomic profiling hematology, molecular MRD testing, qPCR MRD AML, digital PCR MRD detection, NGS MRD detection hematology, minimal residual disease hematology, rapid molecular diagnostics hematology, molecular vs cytogenetic turnaround time, diagnostic testing speed hematology, cytogenetic vs molecular testing comparison, targeted therapy genetic markers, leukemia genetic classification, lymphoma genetic abnormalities, genomic risk stratification hematology, diagnostic workflow hematology, inherited marrow failure genetic testing, whole genome sequencing hematology, exome sequencing hematology, precision medicine hematology, Ask Hematologist, Dr Moustafa Abdou, askhematologist.com